Structural insights into the effects of 2′-5′ linkages on the RNA duplex
Jia Sheng, Li Li, Aaron E. Engelhart, Jianhua Gan, Jiawei Wang, and Jack W. Szostak
Proceedings of the National Academy of Sciences of the United States of America, 2014, 118 (8), pp 3050-3055. doi: 10.1073/pnas.1317799111
Publisher | ResearchGate | PubMed | Google Scholar
Scientific Abstract:
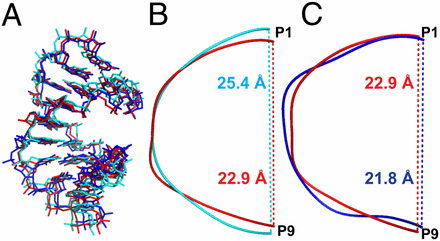
The mixture of 2′-5′ and 3′-5′ linkages generated during the nonenzymatic replication of RNA has long been regarded as a central problem for the origin of the RNA world. However, we recently observed that both a ribozyme and an RNA aptamer retain considerable functionality in the presence of prebiotically plausible levels of linkage heterogeneity. To better understand the RNA structure and function in the presence of backbone linkage heterogeneity, we obtained high-resolution X-ray crystal structures of a native 10-mer RNA duplex (1.32 Å) and two variants: one containing one 2′-5′ linkage per strand (1.55 Å) and one containing three such linkages per strand (1.20 Å). We found that RNA duplexes adjust their local structures to accommodate the perturbation caused by 2′-5′ linkages, with the flanking nucleotides buffering the disruptive effects of the isomeric linkage and resulting in a minimally altered global structure. Although most 2′-linked sugars were in the expected 2′-endo conformation, some were partially or fully in the 3′-endo conformation, suggesting that the energy difference between these conformations was relatively small. Our structural and molecular dynamic studies also provide insight into the diminished thermal and chemical stability of the duplex state associated with the presence of 2′-5′ linkages. Our results contribute to the view that a low level of 2′-5′ substitution would not have been fatal in an early RNA world and may in contrast have been helpful for both the emergence of nonenzymatic RNA replication and the early evolution of functional RNAs.
Lay Abstract:
Modern cells synthesize RNA with sophisticated enzymes that read a “template” strand and synthesize a second strand that is complementary – that is, where there’s an G on the template, there will be a C on the product, and where there’s an A on the template, there will be a U on the product, and so on. The enzymes that do this do so with a great deal of chemical specificity – there are actually two atoms on each nucleotide (A/G/C/U) that form part of the RNA backbone that are chemically almost the same. Despite this, the pocket inside the enzyme that catalyzes the reaction can orient the molecules in exactly the right way so that only the “right” atoms end up in the synthesized RNA strand. We can synthesize RNA in the lab without enzymes, but we aren’t nearly as good at it as the enzymes are. Depending how we set up the experiment, about 10-25% of the time, the “wrong” atom ends up in the RNA backbone, making a kink in the strand of RNA produced (the “2′-5′ linkages” – pronounced “two prime, five prime,” referring to the atoms in the ribose sugar part of RNA – mentioned in the scientific abstract – most RNA found in life has only 3′-5′ linkages). That’s about the best we can do with current technology. For a long time, these kinks have been known to have a few effects on RNA, including destabilizing the double helix structure formed between two strands of RNA, and also being less chemically stable when they’re in a double helix. In this paper, we obtained a structure of RNA with 2′-5′ linkages. To do so, we grew crystals of RNA duplexes that have these changes in the backbone and shot them with X-rays. When you have a single, very pure crystal of a molecule, it will scatter X-rays like a prism in a very specific way that produces a fingerprint. From this fingerprint, you can derive where the atoms are located in the 3-D structure of the molecule. We also examined these structures with computer simulations. We found two important things: 1) Even with 2′-5′ linkages, the RNA double helix looks a lot like the regular one, but we can see some structural features that explain why the double helix might come apart more easily, and 2) the structure we found confirmed what had been thought was true for awhile, but never shown directly (previously, people had suggested this based on computer modeling) – the atoms in a 2′-5′ linkage, when they’re in a double helix, are arranged in just the right orientation so that that linkage is chemically less stable. These results suggest that, even if we didn’t have highly evolved, sophisticated enzymes around, RNA copying likely produced a molecule that looked a lot like modern RNA, and the subtle chemical differences, like 2′-5′ linkages, may not have been as much of a problem as previously thought. This is important, since RNA may have played a key role in the evolution of early life.
Readers might also be interested in another paper on 2′-5′ linkages in RNA we published.