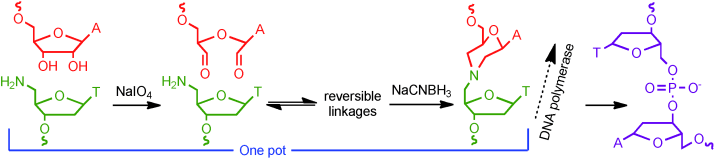
Nonenzymatic Ligation of DNA with a Reversible Step and a Final Linkage that Can Be Used in PCR
Aaron E. Engelhart, Brian J. Cafferty, C. Denise Okafor, Michael C. Chen, Loren Dean Williams, David G. Lynn and Nicholas V. Hud
ChemBioChem, 2012, 13 (8), pp 1121-1124. doi: 10.1002/cbic.201200167
Publisher | ResearchGate | PubMed | Google Scholar
Scientific Abstract
Nonenzymatic DNA ligation chemistries containing a reversible step allow thermodynamic control of product formation, but they are not necessarily compatible with polymerase enzymes. We report a ligation system that uses commercially available reagents, includes a reversible step, and results in a linkage that can function as a template for PCR amplification with accurate sequence transfer.
Lay Abstract
We found a way of joining two pieces of DNA together (“ligation”) with just a few simple chemical reagents. Because our ligation technique doesn’t require enzymes, it’s possible to use it to join together chemically modified pieces of DNA enzymes might reject. Additionally, this technique produces a linkage that, although it’s different than the phosphodiester linkage found in natural DNA, polymerase enzymes that synthesize DNA can move past it while synthesizing another copy of DNA. Many similar ways of performing DNA ligation result in enzymes making a mistake – typically deleting a DNA base or incorporating the wrong base. We also show that one of the enzymes that can read past this linkage produces the correct sequence of DNA – the right base is incorporated on either side of the site where the two strands were joined. Because of this, this method of performing DNA ligation could be useful in biotechnology techniques, potentially including those in live cells.