Primitive Genetic Polymers
Aaron E. Engelhart and Nicholas V. Hud
Cold Spring Harbor Perspectives in Biology, 2010. doi: 10.1101/cshperspect.a002196
Publisher | ResearchGate | PubMed | Google Scholar
Scientific Abstract
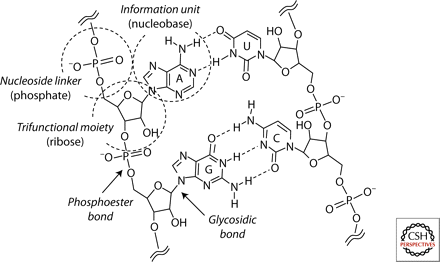
Since the structure of DNA was elucidated more than 50 years ago, Watson-Crick base pairing has been widely speculated to be the likely mode of both information storage and transfer in the earliest genetic polymers. The discovery of catalytic RNA molecules subsequently provided support for the hypothesis that RNA was perhaps even the first polymer of life. However, the de novo synthesis of RNA using only plausible prebiotic chemistry has proven difficult, to say the least. Experimental investigations, made possible by the application of synthetic and physical organic chemistry, have now provided evidence that the nucleobases (A, G, C, and T/U), the trifunctional moiety ([deoxy]ribose), and the linkage chemistry (phosphate esters) of contemporary nucleic acids may be optimally suited for their present roles—a situation that suggests refinement by evolution. Here, we consider studies of variations in these three distinct components of nucleic acids with regard to the question: Is RNA, as is generally acknowledged of DNA, the product of evolution? If so, what chemical and structural features might have been more likely and advantageous for a proto-RNA?
Lay Abstract
In this review, we examine RNA, DNA, and related molecules. Many researchers have considered how the earliest RNA and DNA molecules (or their chemical “cousins”) might have formed, and some have come to the conclusion that the components of these molecules, like many other parts of life, might have been subject to evolution. We separate existing molecules into three parts – the base (A, G, C, and T/U), the “trifunctional moiety” (in DNA and RNA, the sugar molecules deoxyribose and ribose), and the linkage chemistry (in DNA and RNA, phosphate esters) – and examine how these might have evolved, reviewing efforts from others in the field.